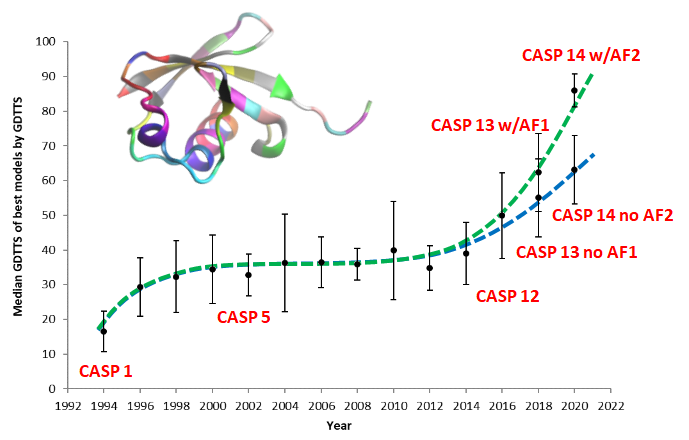
An article covering confident prediction of protein structures, their interactions with other biomolecules, and even protein design, all now at everybody’s reach thanks to the revolution started by Deepmind. Via https://medium.com/advances-in-biological-science/over-one-year-of-alphafold-2-free-for-everyone-to-use-and-of-the-revolution-it-triggered-in-biology-f12cac8c88c6
Contents:
– Introduction
– Democratizing the use of AlphaFold 2 and new adaptations of it
– What’s next
– Protein language models
– Pre-made databases of structural models built with these modern tools
– Unexpected things that AlphaFold 2 seems to be able to do
– Further reads on the topic and on broader structural biology